Upload 32 files
Browse files- .gitattributes +14 -0
- .gitignore +174 -0
- DMDNeuralOperator.py +152 -0
- Experiments/Burgers/DMDNeuralOperator_Experiments_Burgers_Eq.ipynb +3 -0
- Experiments/Burgers/README.md +1 -0
- Experiments/DMDNeuralOperator_Experiments_all.ipynb +3 -0
- Experiments/Heat-Eq/DMDNeuralOperator_Experiments_Heat_Eq.ipynb +0 -0
- Experiments/Heat-Eq/README.md +1 -0
- Experiments/Laplace/DMDNeuralOperator_Experiments_Laplace_Eq.ipynb +0 -0
- Experiments/Laplace/README.md +1 -0
- Experiments/README.md +1 -0
- LICENSE +21 -0
- README.md +88 -3
- doc/Algorithm.png +3 -0
- doc/DMDNeuralOperator_diagram.png +3 -0
- doc/NN_diagram.png +0 -0
- doc/README.md +1 -0
- doc/burgers_eq/loss/Burgers_Eq_Smooth_Loss_DMDNO.png +0 -0
- doc/burgers_eq/test/Burgers_Eq_Smooth_1_DMDNO.png +3 -0
- doc/burgers_eq/test/Burgers_Eq_Smooth_2_DMDNO.png +3 -0
- doc/burgers_eq/test/Burgers_Eq_Smooth_3_DMDNO.png +3 -0
- doc/formula_1.png +0 -0
- doc/formula_2.png +0 -0
- doc/heat_eq/loss/Heat_Eq_Smooth_Loss_DMDNO.png +0 -0
- doc/heat_eq/test/Heat_Eq_Smooth_1_DMDNO.png +3 -0
- doc/heat_eq/test/Heat_Eq_Smooth_2_DMDNO.png +3 -0
- doc/heat_eq/test/Heat_Eq_Smooth_3_DMDNO.png +3 -0
- doc/laplace_eq/loss/Laplace_Eq_Smooth_Loss_DMDNO.png +0 -0
- doc/laplace_eq/test/Laplace_Eq_Smooth_1_DMDNO.png +3 -0
- doc/laplace_eq/test/Laplace_Eq_Smooth_2_DMDNO.png +3 -0
- doc/laplace_eq/test/Laplace_Eq_Smooth_3_DMDNO.png +3 -0
- doc/model_architecture.png +3 -0
- requirements.txt +8 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,17 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
doc/Algorithm.png filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
doc/burgers_eq/test/Burgers_Eq_Smooth_1_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 38 |
+
doc/burgers_eq/test/Burgers_Eq_Smooth_2_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 39 |
+
doc/burgers_eq/test/Burgers_Eq_Smooth_3_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 40 |
+
doc/DMDNeuralOperator_diagram.png filter=lfs diff=lfs merge=lfs -text
|
| 41 |
+
doc/heat_eq/test/Heat_Eq_Smooth_1_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 42 |
+
doc/heat_eq/test/Heat_Eq_Smooth_2_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 43 |
+
doc/heat_eq/test/Heat_Eq_Smooth_3_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 44 |
+
doc/laplace_eq/test/Laplace_Eq_Smooth_1_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 45 |
+
doc/laplace_eq/test/Laplace_Eq_Smooth_2_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 46 |
+
doc/laplace_eq/test/Laplace_Eq_Smooth_3_DMDNO.png filter=lfs diff=lfs merge=lfs -text
|
| 47 |
+
doc/model_architecture.png filter=lfs diff=lfs merge=lfs -text
|
| 48 |
+
Experiments/Burgers/DMDNeuralOperator_Experiments_Burgers_Eq.ipynb filter=lfs diff=lfs merge=lfs -text
|
| 49 |
+
Experiments/DMDNeuralOperator_Experiments_all.ipynb filter=lfs diff=lfs merge=lfs -text
|
.gitignore
ADDED
|
@@ -0,0 +1,174 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Byte-compiled / optimized / DLL files
|
| 2 |
+
__pycache__/
|
| 3 |
+
*.py[cod]
|
| 4 |
+
*$py.class
|
| 5 |
+
|
| 6 |
+
# C extensions
|
| 7 |
+
*.so
|
| 8 |
+
|
| 9 |
+
# Distribution / packaging
|
| 10 |
+
.Python
|
| 11 |
+
build/
|
| 12 |
+
develop-eggs/
|
| 13 |
+
dist/
|
| 14 |
+
downloads/
|
| 15 |
+
eggs/
|
| 16 |
+
.eggs/
|
| 17 |
+
lib/
|
| 18 |
+
lib64/
|
| 19 |
+
parts/
|
| 20 |
+
sdist/
|
| 21 |
+
var/
|
| 22 |
+
wheels/
|
| 23 |
+
share/python-wheels/
|
| 24 |
+
*.egg-info/
|
| 25 |
+
.installed.cfg
|
| 26 |
+
*.egg
|
| 27 |
+
MANIFEST
|
| 28 |
+
|
| 29 |
+
# PyInstaller
|
| 30 |
+
# Usually these files are written by a python script from a template
|
| 31 |
+
# before PyInstaller builds the exe, so as to inject date/other infos into it.
|
| 32 |
+
*.manifest
|
| 33 |
+
*.spec
|
| 34 |
+
|
| 35 |
+
# Installer logs
|
| 36 |
+
pip-log.txt
|
| 37 |
+
pip-delete-this-directory.txt
|
| 38 |
+
|
| 39 |
+
# Unit test / coverage reports
|
| 40 |
+
htmlcov/
|
| 41 |
+
.tox/
|
| 42 |
+
.nox/
|
| 43 |
+
.coverage
|
| 44 |
+
.coverage.*
|
| 45 |
+
.cache
|
| 46 |
+
nosetests.xml
|
| 47 |
+
coverage.xml
|
| 48 |
+
*.cover
|
| 49 |
+
*.py,cover
|
| 50 |
+
.hypothesis/
|
| 51 |
+
.pytest_cache/
|
| 52 |
+
cover/
|
| 53 |
+
|
| 54 |
+
# Translations
|
| 55 |
+
*.mo
|
| 56 |
+
*.pot
|
| 57 |
+
|
| 58 |
+
# Django stuff:
|
| 59 |
+
*.log
|
| 60 |
+
local_settings.py
|
| 61 |
+
db.sqlite3
|
| 62 |
+
db.sqlite3-journal
|
| 63 |
+
|
| 64 |
+
# Flask stuff:
|
| 65 |
+
instance/
|
| 66 |
+
.webassets-cache
|
| 67 |
+
|
| 68 |
+
# Scrapy stuff:
|
| 69 |
+
.scrapy
|
| 70 |
+
|
| 71 |
+
# Sphinx documentation
|
| 72 |
+
docs/_build/
|
| 73 |
+
|
| 74 |
+
# PyBuilder
|
| 75 |
+
.pybuilder/
|
| 76 |
+
target/
|
| 77 |
+
|
| 78 |
+
# Jupyter Notebook
|
| 79 |
+
.ipynb_checkpoints
|
| 80 |
+
|
| 81 |
+
# IPython
|
| 82 |
+
profile_default/
|
| 83 |
+
ipython_config.py
|
| 84 |
+
|
| 85 |
+
# pyenv
|
| 86 |
+
# For a library or package, you might want to ignore these files since the code is
|
| 87 |
+
# intended to run in multiple environments; otherwise, check them in:
|
| 88 |
+
# .python-version
|
| 89 |
+
|
| 90 |
+
# pipenv
|
| 91 |
+
# According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
|
| 92 |
+
# However, in case of collaboration, if having platform-specific dependencies or dependencies
|
| 93 |
+
# having no cross-platform support, pipenv may install dependencies that don't work, or not
|
| 94 |
+
# install all needed dependencies.
|
| 95 |
+
#Pipfile.lock
|
| 96 |
+
|
| 97 |
+
# UV
|
| 98 |
+
# Similar to Pipfile.lock, it is generally recommended to include uv.lock in version control.
|
| 99 |
+
# This is especially recommended for binary packages to ensure reproducibility, and is more
|
| 100 |
+
# commonly ignored for libraries.
|
| 101 |
+
#uv.lock
|
| 102 |
+
|
| 103 |
+
# poetry
|
| 104 |
+
# Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
|
| 105 |
+
# This is especially recommended for binary packages to ensure reproducibility, and is more
|
| 106 |
+
# commonly ignored for libraries.
|
| 107 |
+
# https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
|
| 108 |
+
#poetry.lock
|
| 109 |
+
|
| 110 |
+
# pdm
|
| 111 |
+
# Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control.
|
| 112 |
+
#pdm.lock
|
| 113 |
+
# pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it
|
| 114 |
+
# in version control.
|
| 115 |
+
# https://pdm.fming.dev/latest/usage/project/#working-with-version-control
|
| 116 |
+
.pdm.toml
|
| 117 |
+
.pdm-python
|
| 118 |
+
.pdm-build/
|
| 119 |
+
|
| 120 |
+
# PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm
|
| 121 |
+
__pypackages__/
|
| 122 |
+
|
| 123 |
+
# Celery stuff
|
| 124 |
+
celerybeat-schedule
|
| 125 |
+
celerybeat.pid
|
| 126 |
+
|
| 127 |
+
# SageMath parsed files
|
| 128 |
+
*.sage.py
|
| 129 |
+
|
| 130 |
+
# Environments
|
| 131 |
+
.env
|
| 132 |
+
.venv
|
| 133 |
+
env/
|
| 134 |
+
venv/
|
| 135 |
+
ENV/
|
| 136 |
+
env.bak/
|
| 137 |
+
venv.bak/
|
| 138 |
+
|
| 139 |
+
# Spyder project settings
|
| 140 |
+
.spyderproject
|
| 141 |
+
.spyproject
|
| 142 |
+
|
| 143 |
+
# Rope project settings
|
| 144 |
+
.ropeproject
|
| 145 |
+
|
| 146 |
+
# mkdocs documentation
|
| 147 |
+
/site
|
| 148 |
+
|
| 149 |
+
# mypy
|
| 150 |
+
.mypy_cache/
|
| 151 |
+
.dmypy.json
|
| 152 |
+
dmypy.json
|
| 153 |
+
|
| 154 |
+
# Pyre type checker
|
| 155 |
+
.pyre/
|
| 156 |
+
|
| 157 |
+
# pytype static type analyzer
|
| 158 |
+
.pytype/
|
| 159 |
+
|
| 160 |
+
# Cython debug symbols
|
| 161 |
+
cython_debug/
|
| 162 |
+
|
| 163 |
+
# PyCharm
|
| 164 |
+
# JetBrains specific template is maintained in a separate JetBrains.gitignore that can
|
| 165 |
+
# be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
|
| 166 |
+
# and can be added to the global gitignore or merged into this file. For a more nuclear
|
| 167 |
+
# option (not recommended) you can uncomment the following to ignore the entire idea folder.
|
| 168 |
+
#.idea/
|
| 169 |
+
|
| 170 |
+
# Ruff stuff:
|
| 171 |
+
.ruff_cache/
|
| 172 |
+
|
| 173 |
+
# PyPI configuration file
|
| 174 |
+
.pypirc
|
DMDNeuralOperator.py
ADDED
|
@@ -0,0 +1,152 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import torch
|
| 3 |
+
import torch.nn as nn
|
| 4 |
+
from pydmd import DMD
|
| 5 |
+
|
| 6 |
+
|
| 7 |
+
class DMDProcessor:
|
| 8 |
+
def __init__(self, data: torch.Tensor, rank: int):
|
| 9 |
+
"""Process input data using Dynamic Mode Decomposition.
|
| 10 |
+
|
| 11 |
+
Args:
|
| 12 |
+
data: Input tensor of shape (batch_size, ny, nx)
|
| 13 |
+
rank: Rank for SVD approximation
|
| 14 |
+
"""
|
| 15 |
+
self.data = data
|
| 16 |
+
self.rank = rank
|
| 17 |
+
|
| 18 |
+
def _validate_input(self):
|
| 19 |
+
if self.rank <= 0:
|
| 20 |
+
raise ValueError("Rank must be positive integer")
|
| 21 |
+
|
| 22 |
+
def _compute_dmd(self):
|
| 23 |
+
"""Perform DMD and return reconstructed data."""
|
| 24 |
+
try:
|
| 25 |
+
snapshots = self.data.reshape(self.data.shape[0], -1).T
|
| 26 |
+
dmd = DMD(svd_rank=self.rank)
|
| 27 |
+
dmd.fit(snapshots)
|
| 28 |
+
|
| 29 |
+
if dmd.reconstructed_data is None:
|
| 30 |
+
raise RuntimeError("DMD reconstruction failed")
|
| 31 |
+
|
| 32 |
+
return dmd
|
| 33 |
+
|
| 34 |
+
except Exception as e:
|
| 35 |
+
raise RuntimeError(f"DMD processing failed: {str(e)}")
|
| 36 |
+
|
| 37 |
+
def _calc_energy(self):
|
| 38 |
+
dmd = self._compute_dmd()
|
| 39 |
+
energy = np.cumsum(np.abs(dmd.amplitudes)) / np.sum(np.abs(dmd.amplitudes))
|
| 40 |
+
n_modes = np.argmax(energy > 0.95) + 1
|
| 41 |
+
return n_modes
|
| 42 |
+
|
| 43 |
+
def method(self):
|
| 44 |
+
dmd = self._compute_dmd()
|
| 45 |
+
|
| 46 |
+
modes = [dmd.modes.real[:, i] for i in range(len(dmd.amplitudes))]
|
| 47 |
+
dynamics = [dmd.dynamics.real[i] for i in range(len(dmd.amplitudes))]
|
| 48 |
+
return [modes, dynamics]
|
| 49 |
+
|
| 50 |
+
|
| 51 |
+
class DMDNeuralOperator(nn.Module):
|
| 52 |
+
def __init__(self, branch1_dim, branch_dmd_dim_modes, branch_dmd_dim_dynamics, trunk_dim):
|
| 53 |
+
"""Neural operator with DMD preprocessing.
|
| 54 |
+
|
| 55 |
+
Args:
|
| 56 |
+
branch1_dim: Layer dimensions for primary branch
|
| 57 |
+
branch_dmd_dim_modes: Layer dimensions for DMD modes branch
|
| 58 |
+
branch_dmd_dim_dynamics: Layer dimensions for DMD dynamics branch
|
| 59 |
+
trunk_dims: Layer dimensions for trunk network
|
| 60 |
+
"""
|
| 61 |
+
super(DMDNeuralOperator, self).__init__()
|
| 62 |
+
|
| 63 |
+
modules = []
|
| 64 |
+
for i, h_dim in enumerate(branch1_dim):
|
| 65 |
+
if i == 0:
|
| 66 |
+
in_channels = h_dim
|
| 67 |
+
else:
|
| 68 |
+
modules.append(nn.Sequential(
|
| 69 |
+
nn.Linear(in_channels, h_dim),
|
| 70 |
+
nn.Tanh()
|
| 71 |
+
)
|
| 72 |
+
)
|
| 73 |
+
in_channels = h_dim
|
| 74 |
+
|
| 75 |
+
self._branch_1 = nn.Sequential(*modules)
|
| 76 |
+
|
| 77 |
+
modules = []
|
| 78 |
+
for i, h_dim in enumerate(branch_dmd_dim_modes):
|
| 79 |
+
if i == 0:
|
| 80 |
+
in_channels = h_dim
|
| 81 |
+
else:
|
| 82 |
+
modules.append(nn.Sequential(
|
| 83 |
+
nn.Linear(in_channels, h_dim),
|
| 84 |
+
nn.Tanh()
|
| 85 |
+
)
|
| 86 |
+
)
|
| 87 |
+
in_channels = h_dim
|
| 88 |
+
self._branch_dmd_modes = nn.Sequential(*modules)
|
| 89 |
+
|
| 90 |
+
modules = []
|
| 91 |
+
for i, h_dim in enumerate(branch_dmd_dim_dynamics):
|
| 92 |
+
if i == 0:
|
| 93 |
+
in_channels = h_dim
|
| 94 |
+
else:
|
| 95 |
+
modules.append(nn.Sequential(
|
| 96 |
+
nn.Linear(in_channels, h_dim),
|
| 97 |
+
nn.Tanh()
|
| 98 |
+
)
|
| 99 |
+
)
|
| 100 |
+
in_channels = h_dim
|
| 101 |
+
self._branch_dmd_dynamics = nn.Sequential(*modules)
|
| 102 |
+
|
| 103 |
+
modules = []
|
| 104 |
+
for i, h_dim in enumerate(trunk_dim):
|
| 105 |
+
if i == 0:
|
| 106 |
+
in_channels = h_dim
|
| 107 |
+
else:
|
| 108 |
+
modules.append(nn.Sequential(
|
| 109 |
+
nn.Linear(in_channels, h_dim),
|
| 110 |
+
nn.Tanh()
|
| 111 |
+
)
|
| 112 |
+
)
|
| 113 |
+
in_channels = h_dim
|
| 114 |
+
|
| 115 |
+
self._trunk = nn.Sequential(*modules)
|
| 116 |
+
|
| 117 |
+
self.final_linear = nn.Linear(trunk_dim[-1], 10)
|
| 118 |
+
|
| 119 |
+
def forward(self, f: torch.Tensor, f_dmd_modes: torch.Tensor, f_dmd_dynamics: torch.Tensor, x: torch.Tensor) -> torch.Tensor:
|
| 120 |
+
"""Forward pass.
|
| 121 |
+
|
| 122 |
+
Args:
|
| 123 |
+
f: Input function (batch_size, *spatial_dims)
|
| 124 |
+
x: Evaluation points (num_points, coord_dim)
|
| 125 |
+
|
| 126 |
+
Returns:
|
| 127 |
+
Output tensor (batch_size, num_points)
|
| 128 |
+
"""
|
| 129 |
+
modes, dynamics = f_dmd_modes, f_dmd_dynamics
|
| 130 |
+
|
| 131 |
+
branch_dmd_modes = self._branch_dmd_modes(modes)
|
| 132 |
+
branch_dmd_dynamics = self._branch_dmd_dynamics(dynamics)
|
| 133 |
+
y_branch_dmd = branch_dmd_modes * branch_dmd_dynamics
|
| 134 |
+
|
| 135 |
+
y_branch1 = self._branch_1(f)
|
| 136 |
+
y_br = y_branch1 * y_branch_dmd
|
| 137 |
+
|
| 138 |
+
y_tr = self._trunk(x)
|
| 139 |
+
|
| 140 |
+
y_out = y_br @ y_tr
|
| 141 |
+
|
| 142 |
+
linear_out = nn.Linear(y_out.shape[-1], 10)
|
| 143 |
+
tanh_out = nn.Tanh()
|
| 144 |
+
|
| 145 |
+
y_out = self.final_linear(y_out)
|
| 146 |
+
|
| 147 |
+
return y_out
|
| 148 |
+
|
| 149 |
+
def loss(self, f, f_dmd_modes, f_dmd_dynamics, x, y):
|
| 150 |
+
y_out = self.forward(f, f_dmd_modes, f_dmd_dynamics, x)
|
| 151 |
+
loss = ((y_out - y) ** 2).mean()
|
| 152 |
+
return loss
|
Experiments/Burgers/DMDNeuralOperator_Experiments_Burgers_Eq.ipynb
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0f5174604b5db8deeb1581a9e0aa769a5c2407107423c454de29e28f0a81db23
|
| 3 |
+
size 13637720
|
Experiments/Burgers/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|
Experiments/DMDNeuralOperator_Experiments_all.ipynb
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6026e1170396fb8345cc1b8b013ea66167604bb36b743fb512496c7c79d04868
|
| 3 |
+
size 18274979
|
Experiments/Heat-Eq/DMDNeuralOperator_Experiments_Heat_Eq.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Experiments/Heat-Eq/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|
Experiments/Laplace/DMDNeuralOperator_Experiments_Laplace_Eq.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Experiments/Laplace/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|
Experiments/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|
LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2025 @NekkittAY
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
README.md
CHANGED
|
@@ -1,3 +1,88 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# DMD-Neural-Operator
|
| 2 |
+
|
| 3 |
+
[](https://www.python.org/)
|
| 4 |
+
[](#)
|
| 5 |
+
[](#)
|
| 6 |
+
[](#)
|
| 7 |
+

|
| 8 |
+

|
| 9 |
+
[](https://arxiv.org/abs/2507.01117)
|
| 10 |
+
[](LICENSE)
|
| 11 |
+
|
| 12 |
+
Neural operator architecture that combines Dynamic Mode Decomposition (DMD) with deep learning for solving partial differential equations (PDEs).
|
| 13 |
+
|
| 14 |
+
DMD-Neural-Operator is a novel neural operator architecture that synergistically combines Dynamic Mode Decomposition (DMD) with deep learning to efficiently solve partial differential equations (PDEs). By leveraging DMD for dimensionality reduction and feature extraction, the architecture identifies key modes and system dynamics within PDE solutions. These extracted features are then integrated with neural networks to facilitate operator learning, providing an efficient means of approximating PDE solutions in various parameterized settings. This hybrid approach significantly reduces computational costs compared to traditional methods like FEM, FDM, and FVM, while maintaining high solution reconstruction accuracy, as demonstrated on benchmark problems such as the heat equation, Laplace's equation, and Burgers' equation.
|
| 15 |
+
|
| 16 |
+
* Sakovich, N., Aksenov, D., Pleshakova, E., & Gataullin, S. (2025). A Neural Operator based on Dynamic Mode Decomposition. arXiv preprint arXiv:2507.01117. https://doi.org/10.48550/arXiv.2507.01117
|
| 17 |
+
|
| 18 |
+
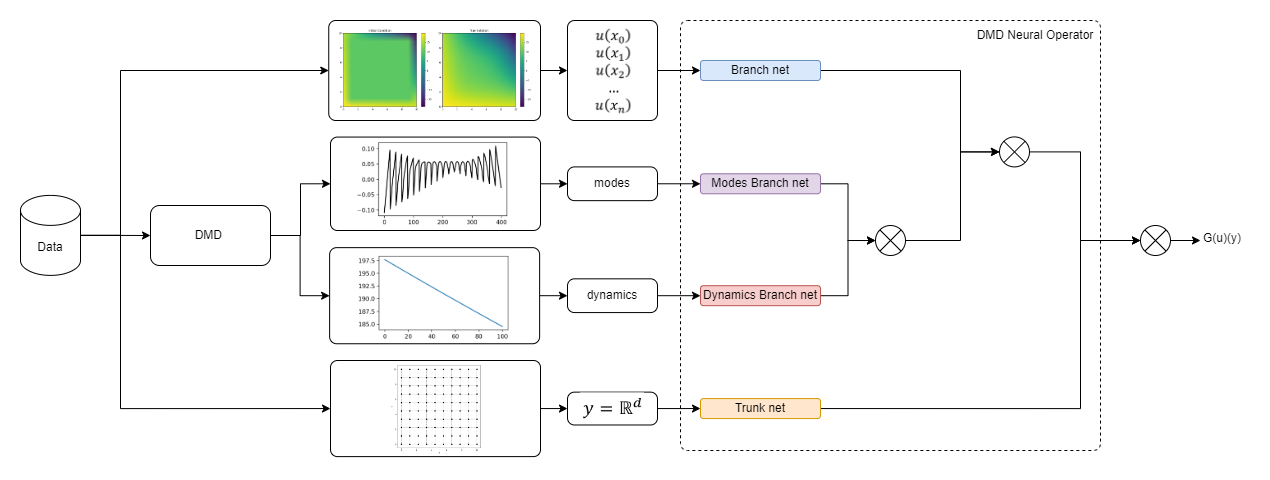
<img width="800px" src="https://github.com/NekkittAY/DMD-Neural-Operator/blob/main/doc/DMDNeuralOperator_diagram.png"/>
|
| 19 |
+
|
| 20 |
+
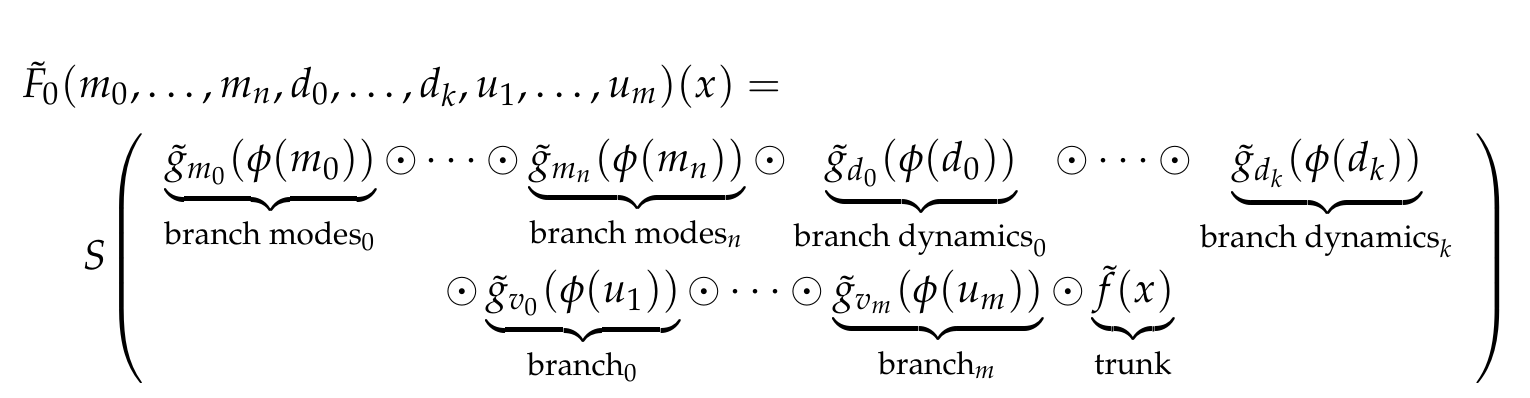
<img width="800px" src="https://github.com/NekkittAY/DMD-Neural-Operator/blob/main/doc/formula_1.png"/>
|
| 21 |
+
<img width="800px" src="https://github.com/NekkittAY/DMD-Neural-Operator/blob/main/doc/formula_2.png"/>
|
| 22 |
+
|
| 23 |
+
## A neural operator using dynamic mode decomposition analysis to approximate the partial differential equations
|
| 24 |
+
|
| 25 |
+
### Abstract
|
| 26 |
+
|
| 27 |
+
Solving partial differential equations (PDEs) for various initial and boundary conditions requires significant computational resources. We propose a neural operator $G_\theta: \mathcal{A} \to \mathcal{U}$, mapping functional spaces, which combines dynamic mode decomposition (DMD) and deep learning for efficient modeling of spatiotemporal processes. The method automatically extracts key modes and system dynamics and uses them to construct predictions, reducing computational costs compared to traditional methods (FEM, FDM, FVM). The approach is demonstrated on the heat equation, Laplace's equation, and Burgers' equation, where it achieves high solution reconstruction accuracy.
|
| 28 |
+
|
| 29 |
+
## Table of Contents
|
| 30 |
+
|
| 31 |
+
- [Overview](#overview)
|
| 32 |
+
- [Technology Stack](#technology-stack)
|
| 33 |
+
- [Features](#features)
|
| 34 |
+
- [Algorithm](#algorithm)
|
| 35 |
+
- [Article](#article)
|
| 36 |
+
|
| 37 |
+
## Overview
|
| 38 |
+
|
| 39 |
+
DMD-Neural-Operator is a hybrid approach that:
|
| 40 |
+
1. Uses DMD for dimensionality reduction and feature extraction from PDE solutions
|
| 41 |
+
2. Combines DMD modes and dynamics with neural networks for operator learning
|
| 42 |
+
3. Provides an efficient way to approximate PDE solutions in parameterized settings
|
| 43 |
+
|
| 44 |
+
## Technology Stack
|
| 45 |
+
|
| 46 |
+
- **Core**: Python 3.8+
|
| 47 |
+
- **Deep Learning**: PyTorch 2.6+
|
| 48 |
+
- **DMD**: PyDMD 2025.4+
|
| 49 |
+
- **Numerical Computing**: NumPy, SciPy
|
| 50 |
+
- **Visualization**: Matplotlib
|
| 51 |
+
- **Development**: tqdm, torchviz
|
| 52 |
+
|
| 53 |
+
## Features
|
| 54 |
+
|
| 55 |
+
- Dimensionality reduction using DMD analysis
|
| 56 |
+
- Neural operator architecture for function space mapping
|
| 57 |
+
- Efficient processing of spatiotemporal data
|
| 58 |
+
- Customizable network architecture with multiple branches
|
| 59 |
+
|
| 60 |
+
## Algorithm
|
| 61 |
+
|
| 62 |
+
<img width="800px" src="https://github.com/NekkittAY/DMD-Neural-Operator/blob/main/doc/Algorithm.png"/>
|
| 63 |
+
|
| 64 |
+
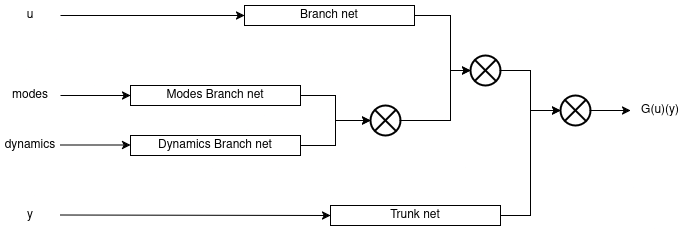
<img width="800px" src="https://github.com/NekkittAY/DMD-Neural-Operator/blob/main/doc/NN_diagram.png"/>
|
| 65 |
+
|
| 66 |
+
|
| 67 |
+
## Article
|
| 68 |
+
```
|
| 69 |
+
@article{sakovich2025neural,
|
| 70 |
+
title={A Neural Operator based on Dynamic Mode Decomposition},
|
| 71 |
+
author={Sakovich, Nikita and Aksenov, Dmitry and Pleshakova, Ekaterina and Gataullin, Sergey},
|
| 72 |
+
journal={arXiv preprint arXiv:2507.01117},
|
| 73 |
+
year={2025}
|
| 74 |
+
}
|
| 75 |
+
```
|
| 76 |
+
|
| 77 |
+
```
|
| 78 |
+
@article{sakovich2025neural,
|
| 79 |
+
title={A neural operator using dynamic mode decomposition analysis to approximate partial differential equations},
|
| 80 |
+
author={Sakovich, Nikita and Aksenov, Dmitry and Pleshakova, Ekaterina and Gataullin, Sergey},
|
| 81 |
+
journal={AIMS Mathematics},
|
| 82 |
+
volume={10},
|
| 83 |
+
number={9},
|
| 84 |
+
pages={22432--22444},
|
| 85 |
+
year={2025}
|
| 86 |
+
}
|
| 87 |
+
```
|
| 88 |
+
|
doc/Algorithm.png
ADDED

|
Git LFS Details
|
doc/DMDNeuralOperator_diagram.png
ADDED
|
Git LFS Details
|
doc/NN_diagram.png
ADDED
|
doc/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|
doc/burgers_eq/loss/Burgers_Eq_Smooth_Loss_DMDNO.png
ADDED

|
doc/burgers_eq/test/Burgers_Eq_Smooth_1_DMDNO.png
ADDED

|
Git LFS Details
|
doc/burgers_eq/test/Burgers_Eq_Smooth_2_DMDNO.png
ADDED

|
Git LFS Details
|
doc/burgers_eq/test/Burgers_Eq_Smooth_3_DMDNO.png
ADDED

|
Git LFS Details
|
doc/formula_1.png
ADDED
|
doc/formula_2.png
ADDED
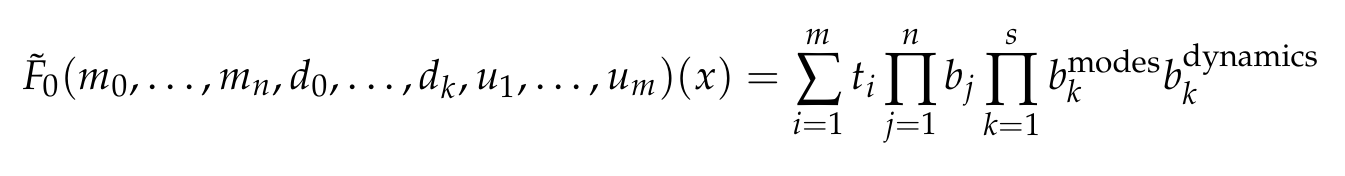
|
doc/heat_eq/loss/Heat_Eq_Smooth_Loss_DMDNO.png
ADDED

|
doc/heat_eq/test/Heat_Eq_Smooth_1_DMDNO.png
ADDED

|
Git LFS Details
|
doc/heat_eq/test/Heat_Eq_Smooth_2_DMDNO.png
ADDED

|
Git LFS Details
|
doc/heat_eq/test/Heat_Eq_Smooth_3_DMDNO.png
ADDED

|
Git LFS Details
|
doc/laplace_eq/loss/Laplace_Eq_Smooth_Loss_DMDNO.png
ADDED

|
doc/laplace_eq/test/Laplace_Eq_Smooth_1_DMDNO.png
ADDED

|
Git LFS Details
|
doc/laplace_eq/test/Laplace_Eq_Smooth_2_DMDNO.png
ADDED

|
Git LFS Details
|
doc/laplace_eq/test/Laplace_Eq_Smooth_3_DMDNO.png
ADDED

|
Git LFS Details
|
doc/model_architecture.png
ADDED
|
Git LFS Details
|
requirements.txt
ADDED
|
@@ -0,0 +1,8 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
numpy~=2.2.4
|
| 2 |
+
scipy~=1.15.2
|
| 3 |
+
matplotlib~=3.10.1
|
| 4 |
+
pandas~=2.2.3
|
| 5 |
+
torch~=2.6.0
|
| 6 |
+
pydmd~=2025.4.1
|
| 7 |
+
tqdm~=4.67.1
|
| 8 |
+
torchviz~=0.0.3
|