ROSIE Model: Training and Evaluation
This repository contains code for training and evaluating the ROSIE model, designed for H&E to multiplex protein prediction in histopathology images.
Overview
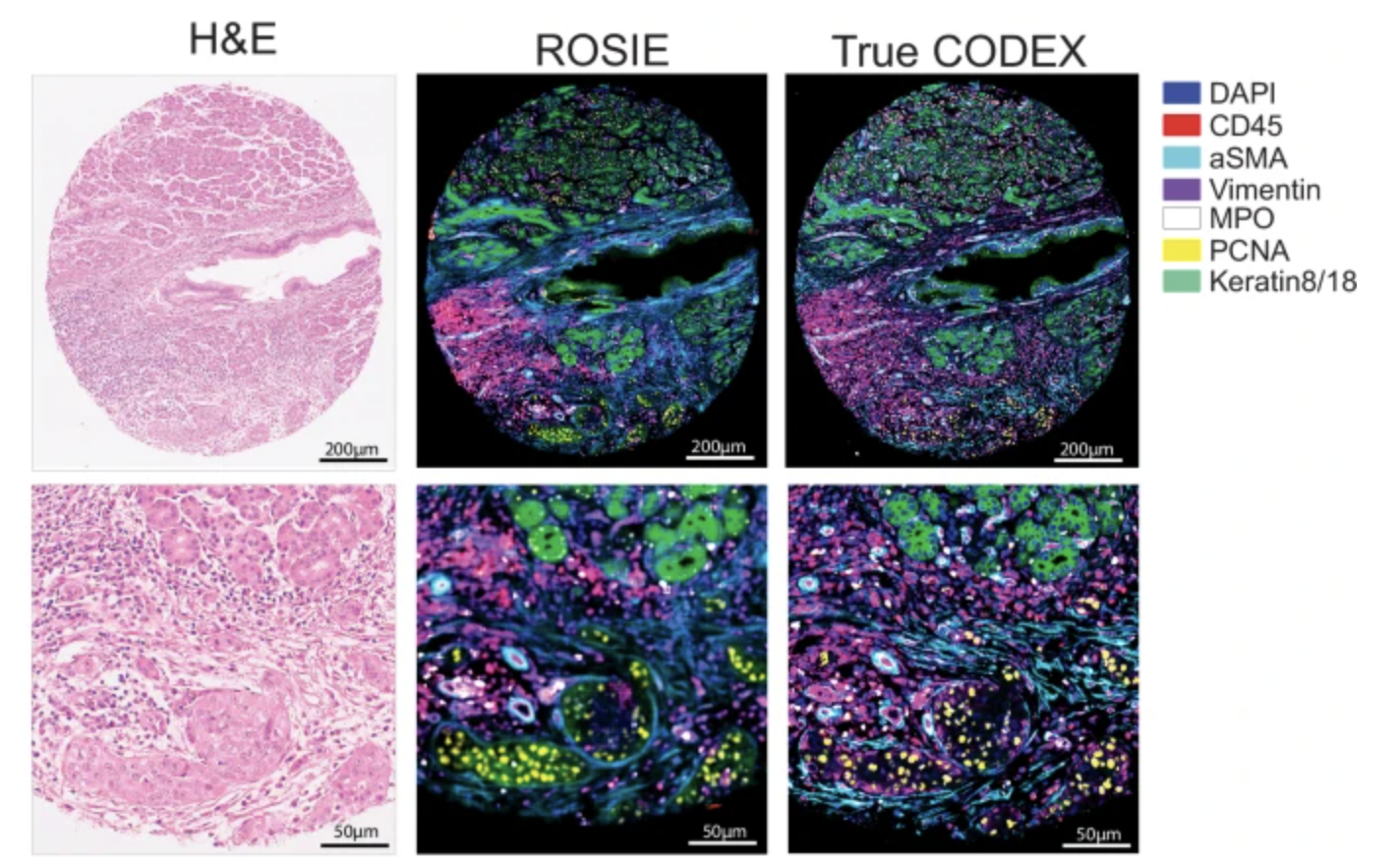
The ROSIE model predicts multiplex protein expression from H&E-stained histopathology images.
The evaluation script processes H&E images (both ZARR and PNG formats) and generates protein expression predictions across 50 channels.
Data and Model Access
To request access to the pretrained model weights, accept the license terms on this page. To request access to the training data, please contact [email protected] and [email protected].
Installation
Install the required dependencies:
pip install -r requirements.txt
Example Usage
Process a single H&E image to generate multiplex protein predictions:
python evaluate.py
--input_dir /path/to/he/images
--output_dir /path/to/output
--model_path /path/to/model.pth
Output:
- A
.tiffimage file with 50 channels. - Channels correspond to the following biomarkers:
DAPI, CD45, CD68, CD14, PD1, FoxP3, CD8, HLA-DR, PanCK, CD3e, CD4, aSMA, CD31, Vimentin, CD45RO, Ki67, CD20, CD11c, Podoplanin, PDL1, GranzymeB, CD38, CD141, CD21, CD163, BCL2, LAG3, EpCAM, CD44, ICOS, GATA3, Gal3, CD39, CD34, TIGIT, ECad, CD40, VISTA, HLA-A, MPO, PCNA, ATM, TP63, IFNg, Keratin8/18, IDO1, CD79a, HLA-E, CollagenIV, CD66
For example:
- Channel 0 → DAPI
- Channel 1 → CD45
- ... and so on.
Postprocessing:
Several postprocessing algorithms are available via the --postprocess_image flag. These adjust channel intensities for human-viewable ranges.
- Note: Min/max intensity depends on protein marker expression in each sample.
- For quantitative analysis, use raw output values.
Directory Structure
Core Scripts
evaluate.py— Evaluation and inference script. Runs inference on H&E images, predicts protein expression, outputs TIFF files.train.py— Training script implementing ConvNeXt-based architecture, patch-based training, augmentation, and evaluation.utils.py— Utility functions for image analysis, ML tasks, metrics, data loading, and visualization.patch_to_cell.py— Converts patch-level predictions into cell-level measurements using segmentation masks.process_exp.py— Processes H&E images into expression predictions and cell-level measurements.reconstruct_codex.py— Reconstructs CODEX images from parquet patch-level expression data.
Note: Most scripts require dependencies and configurations specific to our development environment. However,
evaluate.pycan be run directly with the dependencies listed inrequirements.txt.
Configuration and Data Files
requirements.txt— Python dependencies.Training Datasets.csv— Metadata for training datasets.Antibody Information.xlsx— Antibody and biomarker details for the 50 protein channels.
Citation
If you use this repository in your research, please cite:
Wu, Eric, et al. "ROSIE: AI generation of multiplex immunofluorescence staining from histopathology images." Nature Communications 2025.
👉 ROSIE, Nature Communications 2025
License
This project is licensed under the CC Attribution-NonCommercial 4.0 International.
See the LICENSE file for details.